Pandas
Understanding pandas errors
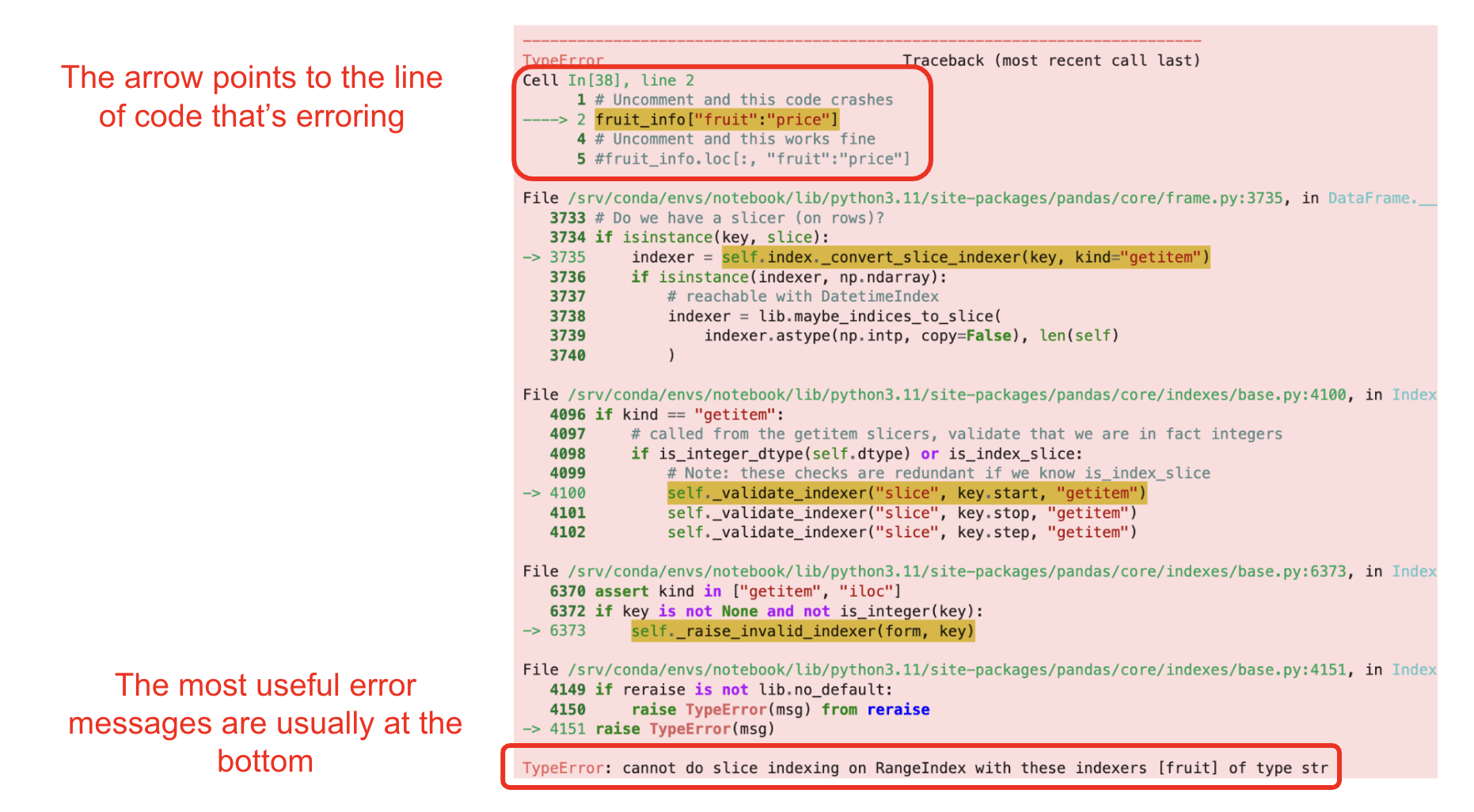
pandas errors can look red, scary, and very long. Fortunately, we don’t need to understand the entire thing! The most important parts of an error message are at the top, which tells you which line of code is causing the issue, and at the bottom, which tells you exactly what the error message is.
This note is (mostly) structured around the error messages that show up at the bottom.
My code is taking a really long time to run
It is normal for a cell to take a few seconds – sometimes a few minutes – to run. If it’s is taking too long, however, you have several options:
- Try restarting the kernel. Sometimes, Datahub glitches or lags, causing the code to run slower than expected. Restarting the kernel should fix this problem, but if the cell is still taking a while to run, it is likely a problem with your code.
- Scrutinize your code. Am I using too many for loops? Is there a repeated operation that I can substitute with a
pandasfunction?
Why is it generally better avoid using loops or list comprehensions when possible?
In one word: performance. NumPy and pandas functions are optimized to handle large amounts of data in an efficient manner. Even for simple operations, like the elementwise addition of two arrays, NumPy arrays are much faster and scale better (feel free to experiment with this yourself using %%time). This is why we encourage you to vectorize your code (ie. using NumPy arrays, Series, or DataFrames instead of Python lists) and use in-built NumPy/pandas functions wherever possible.
KeyErrors
KeyError: 'column_name'
This error usually happens when we have a DataFame called df, and we’re trying to do an operation on a column 'column_name' that does not exist. If you encounter this error, double check that you’re operating on the right column. It might be a good idea to display df and see what it looks like. You could also call df.columns to list all the columns in df.
TypeErrors
TypeError: '___' object is not callable
This often happens when you use a default keyword (like str, list, range, bool, sum, or max) as a variable name, for instance:
sum = 1 + 2 + 3These errors can be tricky because they don’t error on their own but cause problems when we try to use the name sum (for example) later on in the notebook.
To fix the issue, identify any such lines of code (Ctrl+F on “sum =” for example), change your variable names to be something more informative, and restart your notebook.
Python keywords like str and list appear in green text, so be on the lookout if any of your variable names appear in green!
TypeError: could not convert string to a float
This error often occurs when we try to do math operations (ie. sum, average, min, max) on a DataFrame column or Series that contains strings instead of numbers (note that we can do math operations with booleans; Python treats True as 1 and False as 0).
Double check that the column you’re interested in is a numerical type (int, float, or double). If it looks like a number, but you’re still getting this error, you can use .astype(...) (documentation) to change the datatype of a DataFrame or Series.
TypeError: Could not convert <string> to numeric
Related to the above (but distinct), you may run into this error when performing a numeric aggregation function (like mean or sum functions that take integer arguments) after doing a groupby operation on a DataFrame with non-numeric columns.
Working with the elections dataset for example,
elections.groupby('Year').agg('mean')would error because pandas cannot compute the mean of the names of presidents. There are three ways to get around this:
- Select only the numeric columns you are interested in before applying the aggregation function. In the above case, both
elections.groupby('Year')['Popular Vote']orelections['Popular vote'].groupby('Year')would work. - Setting the
numeric_onlyargument toTruein the.aggcall, thereby applying the aggregation function only to numeric columns. For example,elections.groupby('Year').agg('mean', numeric_only=True). - Passing in a dictionary to
.aggwhere you specify the column you are applying a particular aggregation function to. Continuing the same example, this looks likeelections.groupby('Year').agg({'Popular vote' : 'mean').
TypeError: 'NoneType' object is not subscriptable / AttributeError: 'NoneType' object has no attribute 'shape'
This usually occurs as you assign a None value to a variable, then try to either index into or access some attribute of that variable. For Python functions like append and extend, you do not need to do any variable assignment because they mutate the variable directly and return None. Assigning None tends to happen as a result of code like:
some_list = some_list.append(element)In contrast, an operation like np.append does not mutate the variable in place and, instead, returns a copy. In these cases, (re)assignment is necessary:
some_array = np.append(some_array, element)TypeError: 'int'/'float' object is not subscriptable
This occurs when you try and index into an integer or other numeric Python data type. It can be confusing to debug amidst a muddle of code, but you can use the error message to identify which variable is causing this error. Using type(var_name) to check the data type of the variable in question can be a good starting point.
IndexErrors
IndexError: invalid index to scalar variable.
This error is similar to the last TypeError in the previous section. However, it is slightly different in that scalar variables come up in the context of NumPy data types which have slightly different attributes.
For a concrete example, if you defined
numpy_arr = np.array([1])and indexed into it twice (numpy_arr[0][0]), you would get the above error. Unlike a Python integer whose type is int, type(numpy_arr[0]) returns the NumPy version of an integer, numpy.int64. Additionally, you can check the data type by accessing the .dtype attribute of NumPy array (numpy_arr.dtype) or scalar variable (numpy_arr[0].dtype).
IndexError: index _ is out of bounds for axis _ with size _
This error usually happens when you try to index a value that’s greater than the size of the array/list/DataFrame/Series. For example,
some_list = [2, 4, 6, 8]some_list has a length of 4. Trying some_list[6] will error because index 6 is greater than the length of the array. Note that some_list[4] will also cause an IndexError because Python and pandas uses zero indexing, which means that the first element has index 0, the second element has index 1, etc.; some_list[4] would grab the fifth element, which is impossible when the list only has 4 elements.
ValueErrors
ValueError: Truth value of a Series is ambiguous
This error could occur when you apply Python logical operators (or, and, not), which only operate on a single boolean values, to NumPy arrays or Series objects, which can contain multiple values. The fix is to use bitwise operators |, &, ~ , respectively, to allow for element-wise comparisons between values in arrays or Series.
Alternatively, these errors could emerge due to overwriting Python keywords like bool and sum that may be used in the autograder tests, similar to what’s described here. You should follow a similar procedure of identifying the line of code erroring, checking if you’ve overwritten any Python keywords using Ctrl+F, and renaming those variables to something more informative before restarting your kernel and running the erroring tests again.
ValueError: Can only compare identically-labeled Series objects
As the message would suggest, this error occurs when comparing two Series objects that have different lengths. You can double check the lengths of the Series using len(series_name) or series_name.size.
ValueError: -1 is not in range / KeyError: -1
This error occurs when you try and index into a Series or DataFrame as you would a Python list. Unlike a list where passing an index of -1 gives the last element, pandas interprets df[-1] as an attempt to find the row corresponding to index -1 (that is, df.loc[-1]). If your intention is to pick out the last row in df, consider using integer-position based indexing by doing df.iloc[-1]. In general, to avoid ambiguity in these cases, it is also good practice to write out both the row and column indices you want with df.iloc[-1, :].
