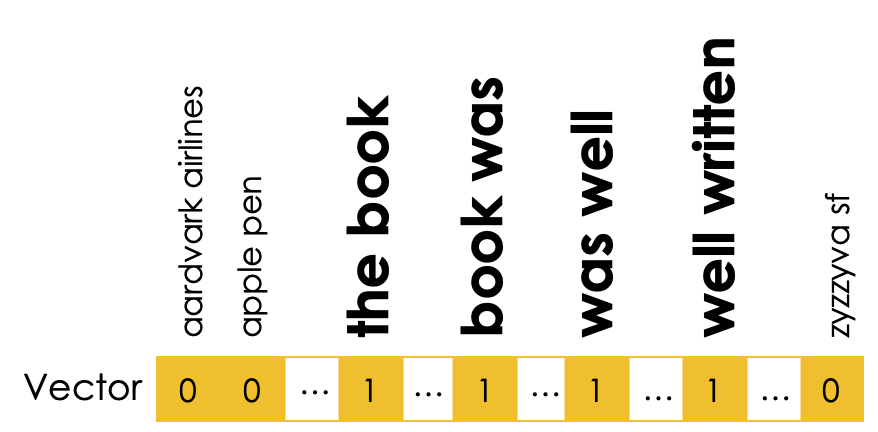
Applying Domain Knowledge¶
The displacement of an engine is defined as the product of the volume of each cylinder and number of cylinders. However, not all cylinders fire at the same time (at least in a functioning engine) so the fuel economy might be more closely related to the volume of any one cylinder.

We can use this "domain knowledge" to compute a new feature encoding the volume per cylinder by taking the ratio of displacement and cylinders.