import numpy as np
import pandas as pd
# Plotly plotting support
import plotly.plotly as py
# import plotly.offline as py
# py.init_notebook_mode()
# import cufflinks as cf
# cf.go_offline() # required to use plotly offline (no account required).
import plotly.graph_objs as go
import plotly.figure_factory as ff
# Make the notebook deterministic
np.random.seed(42)
Cross Validation and the Bias Variance Tradeoff¶
This notebook which accompanies the lecture on the Bias Variance Tradeoff and Regularization.
Notebook created by Joseph E. Gonzalez for DS100.
Toy Dataset¶
As with the previous lectures we will continue to use an easy to visualize synthetic dataset.
np.random.seed(42)
n = 50
sigma = 10
X = np.linspace(-10, 10, n)
X = np.sort(X)
Y = 2. * X + 10. + sigma * np.random.randn(n) + 20*np.sin(X) + 0.8*(X)**2
X = X/5
data_points = go.Scatter(name="data", x=X, y=Y, mode='markers')
py.iplot([data_points])
Making a train Test Split¶
It is always a good habbit to split data into training and test sets.
from sklearn.model_selection import train_test_split
X_tr, X_te, Y_tr, Y_te = train_test_split(X, Y, test_size=0.25, random_state=42)
train_points = go.Scatter(name="Train Data",
x=X_tr, y=Y_tr, mode='markers', marker=dict(color="blue", symbol="o"))
test_points = go.Scatter(name="Test Data",
x=X_te, y=Y_te, mode='markers', marker=dict(color="red", symbol="x"))
py.iplot([train_points, test_points])
Polynomial Features:¶
In the following we work through a basic bias variance tradeoff for different numbers of polynomial features. To make things a bit more interesting we will also add an additional sine feature:
$$ \large \Phi_d(x) = \left[\sin(\omega x), x, x^2, \ldots, x^d \right] $$with the appropriate $\omega = 5$ value determined from prior knowledge (because we created the data).
We use the following Python function to generate a $\Phi$:
def poly_phi(k):
return lambda X: np.array([np.sin(X*5)] + [X ** i for i in range(1, k+1)]).T
We will consider the following basis sizes:
basis_sizes = [1, 2, 5, 8, 16, 24, 32]
Create a $\Phi$ feature matrix for each number of basis:
Phis_tr = { k: poly_phi(k)(X_tr) for k in basis_sizes }
Train a model for each number of feature
from sklearn import linear_model
models = {}
for k in Phis_tr:
model = linear_model.LinearRegression()
model.fit(Phis_tr[k], Y_tr)
models[k] = model
Visualize the various models¶
The following code is a bit complicated but essentially makes an interactive visualization of the various models.
# Make the x values where plot points will be generated
X_plt = np.linspace(np.min(X)-1, np.max(X)+1, 200)
# Generate the Plotly line objects by predicting the value at each X_plt
lines = []
for k in sorted(models.keys()):
ytmp = models[k].predict(poly_phi(k)(X_plt))
# Plotting software fails with large numbers
ytmp[ytmp > 500] = 500
ytmp[ytmp < -500] = -500
lines.append(
go.Scatter(name="degree "+ str(k), x=X_plt,
y = ytmp,visible=False))
# Construct steps for the interactive slider
lines[0].visible=True
steps = []
for i in range(len(lines)):
step = dict(
label = lines[i]['name'],
method = 'restyle',
args = ['visible', [False] * (len(lines)+1)],
)
step['args'][1][0] = True
step['args'][1][i+1] = True # Toggle i'th trace to "visible"
steps.append(step)
# Build the slider object
sliders = [dict(active = 0, pad = {"t": 20}, steps = steps)]
# render the plot
layout = go.Layout(xaxis=dict(range=[np.min(X_plt), np.max(X_plt)]),
yaxis=dict(range=[np.min(Y) -5 , np.max(Y)+5]),
sliders=sliders)
py.iplot(go.Figure(data = [train_points] + lines, layout=layout), filename="poly_curve_comparison")
Cross Validation¶
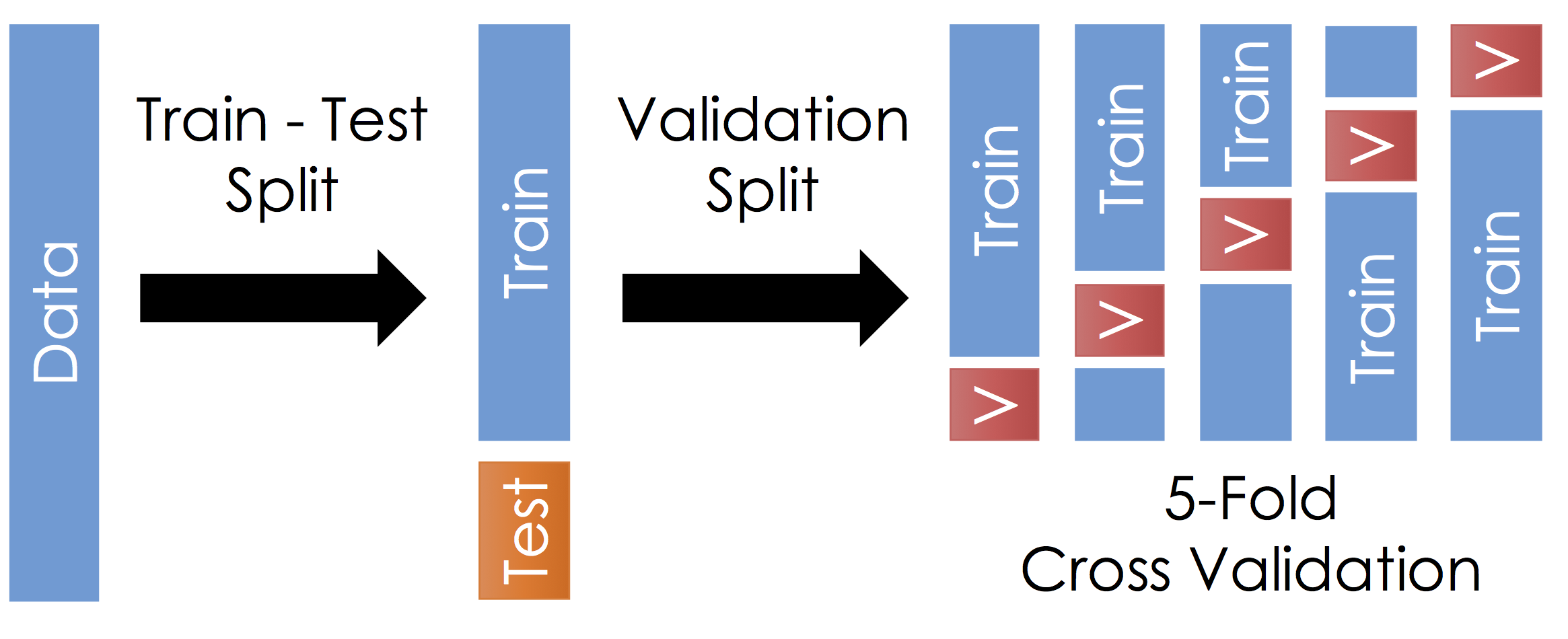
With the remaining training data:
- Split the remaining training dataset k-ways as in the Figure above. The figure uses 5-Fold which is standard. You should split the data in the same way you constructed the test set (this is typically randomly)
- For each split train the model on the training fraction and then compute the error (RMSE) on the validation fraction.
- Average the error across each validation fraction to estimate the test error.
Define the error Function¶
We will continue to use the mean squared error but this time rather than define it ourselves we will import it from scikit-learn.
from sklearn.metrics import mean_squared_error
In the following we compute the cross validated RMSE for different numbers of polynomial basis values
from sklearn.model_selection import KFold
kfold_splits = 5
kfold = KFold(kfold_splits, shuffle=True, random_state=42)
mse_scores = []
poly_degrees = np.arange(1,16)
for k in poly_degrees:
Phi = poly_phi(k)(X_tr)
# One step in k-fold cross validation
def score_model(train_index, test_index):
model = linear_model.LinearRegression()
model.fit(Phi[train_index,], Y_tr[train_index])
return mean_squared_error(Y_tr[test_index], model.predict(Phi[test_index,]))
score = np.mean([score_model(tr_ind, te_ind)
for (tr_ind, te_ind) in kfold.split(Phi)])
mse_scores.append(score)
rmse_scores = np.sqrt(np.array(mse_scores))
py.iplot(
go.Figure(
data=[go.Scatter(name="CV Curve", x=poly_degrees, y=rmse_scores),
go.Scatter(name="Optimum", x=[poly_degrees[np.argmin(rmse_scores)]], y=[np.min(rmse_scores)],
mode="markers", marker=dict(color="red", size=10))
],
layout=go.Layout(xaxis=dict(title="Degree"), yaxis=dict(title="CV RMSE"))),
filename="basis_function_cv_curve")
Questions?¶
- Is 2 a reasonable answer given the data? (Look at the earlier plot.)
- Does this depend on the order in which we consider basis?
Variance¶
What happens if we repeat the training procedure with different versions of our trainin data?
How will the models change as we inrease the degree of our polynomial?
Here we use cross validation to simulate multiple train and test splits by splitting the training data into train and validation datasets. We then visualize each of the corresponding models.
from sklearn.model_selection import KFold
# Construct a KFold object which will define random index splits
kfold_splits = 5
kfold = KFold(kfold_splits, shuffle=True, random_state=42)
# Construct the lines
lines = []
# Make the test lines
X_plt = np.linspace(np.min(X)-1, np.max(X)+1, 200)
# for each train validation split (we ignore the validation data)
for k in basis_sizes:
for (tr_ind, val_ind) in kfold.split(X_tr):
# Construct the Phi matrices for each basis size
Phi = poly_phi(k)(X_tr[tr_ind])
# Fit the model
model = linear_model.LinearRegression()
model.fit(Phi, Y_tr[tr_ind])
# Make predictions at the plotting points
ytmp = model.predict(poly_phi(k)(X_plt))
# Plotly has a bug and fails with large numbers
ytmp[ytmp > 500] = 500
ytmp[ytmp < -500] = -500
line = go.Scatter(name="degree "+ str(k), visible=False,
x=X_plt, y=ytmp)
lines.append(line)
for l in lines[0:kfold_splits]:
l.visible=True
steps = []
for i in range(len(basis_sizes)):
step = dict(
label = "Degree " + str(basis_sizes[i]) ,
method = 'restyle',
args = ['visible', [False] * (len(lines)+1)]
)
step['args'][1][-1] = True
for u in range(i*kfold_splits, (i+1)*kfold_splits):
step['args'][1][u] = True # Toggle i'th trace to "visible"
steps.append(step)
sliders = [dict(
active = 0,
pad = {"t": 20},
steps = steps
)]
layout = go.Layout(xaxis=dict(range=[np.min(X_plt), np.max(X_plt)]),
yaxis=dict(range=[np.min(Y) -5 , np.max(Y) + 5]),
sliders=sliders,
showlegend=False)
py.iplot(go.Figure(data = lines + [train_points], layout=layout), filename="cv_bv_slider")